
本文共 7911 字,大约阅读时间需要 26 分钟。

00. 写在前面
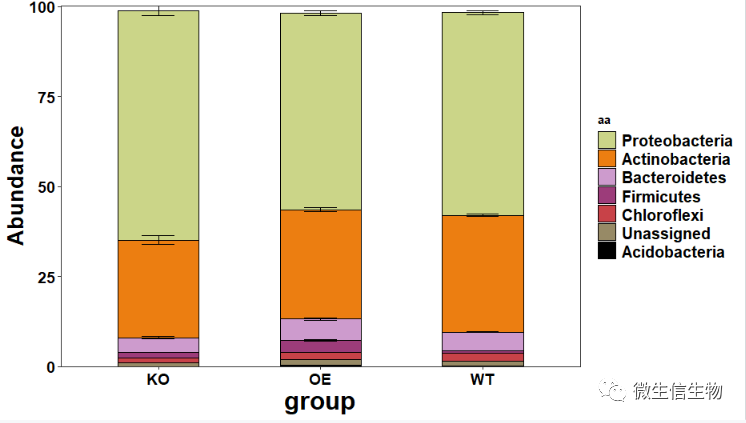
堆叠柱状图用于表征高维数据,例如微生物群落,代谢物组成是一种常见的图表类型,所以使用的次数也非常多,理所应当被封装成函数,一键绘制。
冲击图其实添加了堆叠柱状图之间的连线,这对于柱子之间的横向比较会更加容易一些,同堆叠柱状图互为替代。
堆叠柱状图在R语言中实现似乎不难,但是有许多的细节需要把握:整条柱子的子柱块排序问题,是否在柱子上添加标签,过滤低丰度柱子等在我们这条代码中都会解决。甚至前一段时间大家需要在堆叠柱状图上添加误差线,也都在这条代码里的。
01. 一条函数和参数解释
result = barplot(otu = "./otutab.txt",tax = "./taxonomy.txt",map = "./metadata.tsv",j = "Phylum",k= 0.01,rep = 6,axis_ord = "KO-OE-WT",label = FALSE,sd = TRUE)

otu = "./otutab.txt" :OTU表格
tax = "./taxonomy.txt" :物种注释文件
map = "./metadata.tsv":样本分组文件
j = "Phylum" :设置按照门为分类水平合并OTU,当然可以选择不同的分类水平;j = "Class";j = "Order";j = "Family";j = "Genus";甚至是OTU,如果你不嫌图例多的话。
k= 0.01:按照丰度过滤掉第丰度的物种,减少图例信息。
rep = 6 :测定每个处理重复数量。
axis_ord = "KO-OE-WT" :柱子在x轴额排布顺序。
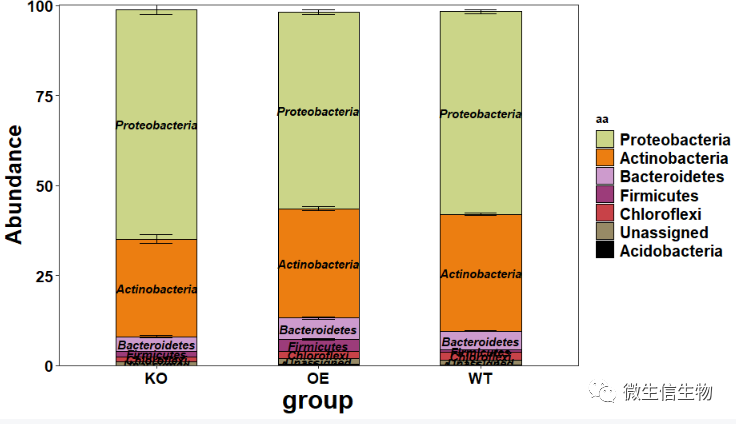
label = FALSE:是否在柱子上添加标签。
sd = TRUE :是否对堆叠柱状图添加标准
02. 处理本地输出图片外我们还可以通过result提取图片
我设置函数返回图形结果的目的是方便大家根据自己需求修改颜色等图形属性。
#提取堆叠柱状图 p = result[[1]] p #提取冲击图 p = result[[2]] p

3. 附:主函数
注意一下几个包,大家都看看装上没有,务必都安装后在运行,方可成功。
library(phyloseq) library(tidyverse) library(vegan) library(reshape2) library("plyr") library(ggalluvial) library(ggplot2) barplot = function(otu = "./otutab.txt",tax = "./taxonomy.txt",map = "./metadata.tsv",j = "Phylum",k= 0.01,rep = 6,axis_ord = "KO-OE-WT",label = TRUE){ library(phyloseq) library(tidyverse) library(vegan) library(reshape2) library("plyr") library(ggalluvial) library(ggplot2) axis_order = strsplit(basename(axis_order), "-")[[1]] #导入otu表格 otu = read.delim(otu,row.names = 1) head(otu) otu = as.matrix(otu) str(otu) #导入注释文件 tax = read.delim(tax,row.names = 1) head(tax) tax = as.matrix(tax) # taxa_names(tax) #导入分组文件 map = read.delim(map,row.names = 1) head(map) # #导入进化树 # tree = read.tree("./otus.tree") # tree ps taxa_are_rows=TRUE), sample_data(map) , tax_table(tax) # phy_tree(tree) ) ps ps1 = ps i = ps1 colnames(tax_table(ps1)) ##这里我们过滤一定阈值的otu,会出现最后堆叠柱状图总体丰度高于100%的情况,这是合理的 ###########绘制不同分类等级的柱状图 Taxonomies % tax_glom(taxrank = j) %>% # agglomerate at Class level Class transform_sample_counts(function(x) {x/sum(x)} )%>%# Transform to rel. abundance psmelt() %>% # Melt to long format filter(Abundance >= k) %>% # Filter out low abundance taxa arrange(Phylum) # head(Taxonomies) # dim(Taxonomies) colbar 1] colors = colorRampPalette(c( "#CBD588", "#599861", "orange","#DA5724", "#508578", "#CD9BCD", "#AD6F3B", "#673770","#D14285", "#652926", "#C84248", "#8569D5", "#5E738F","#D1A33D", "#8A7C64","black"))(colbar) # mi = colorRampPalette(c( "#CBD588", "#599861", "orange","#DA5724", "#508578", "#CD9BCD", # "#AD6F3B", "#673770","#D14285", "#652926", "#C84248", # "#8569D5", "#5E738F","#D1A33D", "#8A7C64","black"))(colbar) # # Taxonomies$Abundance = Taxonomies$Abundance * 100 Taxonomies$Abundance = Taxonomies$Abundance/rep # head(Taxonomies) #按照分组求均值 colnames(Taxonomies) "aa",colnames(Taxonomies)) by_cyl zhnagxu2 = dplyr :: summarise(by_cyl, sum(Abundance)) #colnames(zhnagxu2) = c("group", j,"Abundance") # head(zhnagxu2) ##确定因子,这里通过求和按照从小到大的顺序得到因子 ##长变宽 # head(Taxonomies) Taxonomies2 = dcast(Taxonomies,aa ~ Sample,value.var = "Abundance") head(Taxonomies2) Taxonomies2[is.na(Taxonomies2)] 0 aa = Taxonomies2 # head(aa) n = ncol(aa) #增加一行,为整列的均值,计算每一列的均值,2就是表示列 aa[n+1]=apply(aa[,c(2:ncol(aa))],1,sum) colnames(aa)[n+1] "allsum") # str(aa) bb # head(bb) bb = bb[c(1,ncol(bb))] cc # head(cc) ##使用这个属的因子对下面数据进行排序 head(zhnagxu2) colnames(zhnagxu2) "group","aa","Abundance") zhnagxu2$aa = factor(zhnagxu2$aa,order = T,levels = cc$aa) zhnagxu3 = plyr::arrange(zhnagxu2,desc(aa)) # head(zhnagxu3) ##制作标签坐标,标签位于顶端 # Taxonomies_x = ddply(zhnagxu3,"group", transform, label_y = cumsum(Abundance)) # head(Taxonomies_x ) #标签位于中部 Taxonomies_x = ddply(zhnagxu3,"group", transform, label_y = cumsum(Abundance) - 0.5*Abundance) head(Taxonomies_x,20 ) Taxonomies_x$label = Taxonomies_x$aa #使用循环将堆叠柱状图柱子比较窄的别写标签,仅仅宽柱子写上标签 for(i in 1:nrow(Taxonomies_x)){ if(Taxonomies_x[i,3] > 3){ Taxonomies_x[i,5] = Taxonomies_x[i,5] }else{ Taxonomies_x[i,5] = NA } } ##普通柱状图 p4 x = group, y = Abundance, fill = aa, order = aa)) + geom_bar(stat = "identity",width = 0.5,color = "black") + scale_fill_manual(values = colors) + theme(axis.title.x = element_blank()) + theme(legend.text=element_text(size=6)) + scale_y_continuous(name = "Abundance (%)")+ scale_x_discrete(limits = axis_order) if (label == TRUE) { p4 = p4 + geom_text(aes(y = label_y, label = label ),size = 4,fontface = "bold.italic") } # print(p4) # install.packages("ggalluvial") p4 = p4+theme_bw()+ scale_y_continuous(expand = c(0,0))+ theme( panel.grid.major=element_blank(), panel.grid.minor=element_blank(), text=element_text(face = "bold"), plot.title = element_text(vjust = -8.5,hjust = 0.1), axis.title.y =element_text(size = 20,face = "bold",colour = "black"), axis.title.x =element_text(size = 24,face = "bold",colour = "black"), axis.text = element_text(size = 20,face = "bold"), axis.text.x = element_text(colour = "black",size = 14,), axis.text.y = element_text(colour = "black",size = 14), legend.text = element_text(size = 15) #legend.position = "none"#是否删除图例 ) p4 FileName1 "./a2_",j,k,"_bar",".pdf", sep = "") ggsave(FileName1, p4, width = 12, height =8 ) ##柱状图冲击图 #stratum定义堆叠柱状图柱子内容,以weight定义柱子长度,alluvium定义连线 head(Taxonomies_x ) cs = Taxonomies_x $aa # head(cs) # as.factor(Taxonomies_x $Genus) # cs = as.character(Taxonomies_x $Genus) # cs1 = as.factor(cs) cs1 = cs #提取真正的因子的数量 lengthfactor = length(levels(cs1)) #提取每个因子对应的数量 cs3 = summary (as.factor(cs1)) cs4 = as.data.frame(cs3) cs4$id = row.names(cs4) #对因子进行排序 df_arrange #对Taxonomies_x 对应的列进行排序 Taxonomies_x1 head(Taxonomies_x1) #构建flow的映射列Taxonomies_x Taxonomies_x1$ID = factor(rep(c(1:lengthfactor), cs4$cs3)) #colour = "black",size = 2,,aes(color = "black",size = 0.8) p3 = ggplot(Taxonomies_x1, aes(x = group, stratum = aa, alluvium = ID, weight = Abundance, fill = aa, label = aa)) + geom_flow(stat = "alluvium", lode.guidance = "rightleft", color = "black",size = 0.2,width = 0.3,alpha = .2) + geom_bar(width = 0.45)+ geom_stratum(width = 0.45,size = 0.2) + #geom_text(stat = "stratum", size = 3,family="Times New Roman",fontface = "bold.italic") + #theme(legend.position = "none") + scale_fill_manual(values = colors)+ #ggtitle("fow_plot")+ scale_x_discrete(limits = axis_order)+ geom_text(aes(y = label_y, label = label ),size = 4,fontface = "bold.italic")+ labs(x="group", y="Relative abundancce (%)", ) # p3 if (label == TRUE) { p3 = p3 + geom_text(aes(y = label_y, label = label ),size = 4,fontface = "bold.italic") } p3 =p3+theme_bw()+ scale_y_continuous(expand = c(0,0))+ #geom_hline(aes(yintercept=0), colour="black", linetype=2) + #geom_vline(aes(xintercept=0), colour="black", linetype="dashed") + #scale_fill_manual(values = mi, guide = guide_legend(title = NULL))+ theme( panel.grid.major=element_blank(), panel.grid.minor=element_blank(), text=element_text(face = "bold"), plot.title = element_text(vjust = -8.5,hjust = 0.1), axis.title.y =element_text(size = 20,face = "bold",colour = "black"), axis.title.x =element_text(size = 24,face = "bold",colour = "black"), axis.text = element_text(size = 20,face = "bold"), axis.text.x = element_text(colour = "black",size = 14), axis.text.y = element_text(colour = "black",size = 14), legend.text = element_text(size = 15,face = "bold.italic") #legend.position = "none"#是否删除图例 ) p3 FileName2 ./a2_",j,k,"_bar_flow",".pdf", sep = "") ggsave(FileName2, p3, width = 12, height =8) return(list(p4,p3)) } 

转载地址:https://blog.csdn.net/weixin_34214135/article/details/112594462 如侵犯您的版权,请留言回复原文章的地址,我们会给您删除此文章,给您带来不便请您谅解!
发表评论
最新留言
关于作者
